Small Mammal Diversity & Evolution
Understanding patterns that generate species diversity within a complex island system
Indo-Australian Archipelago is among the most complex island system on earth. Some islands, such as Sumatra, Java, and Borneo, were continuous landmass during the Pleistocene low see level, called Sunda Shelf. Sulawesi was separate islands. Other islands never been connected to other landmasses, such as the Moluccas and Lesser Sunda islands. Within the islands, isolated mountains present more opportunities for speciation. We work on deciphering small mammal diversity and evolution in this complex island system, with particular focus on Sulawesi and the Sunda Shelf.


Project 1
In situ diversification of Hill Rats (Bunomys) on Sulawesi
The hill-rat species group, also referred as the Bunomys Clade, is the most diverse endemic rodent group on Sulawesi. From a single known colonizing ancestor, the group has diversified into 18+ species. Some species occur throughout the island. The other species are restricted to a single area of endemism or only known from a single mountain. This project is the main focus of my PhD. We hope to understand how complex geological history of Sulawesi shaped the diversity of the group. Our understanding of this process will help us to understand the evolution of small mammals in complex island systems. It will also provide an improved understanding of species distribution to inform conservation efforts on Sulawesi.
Publications:
Handika et. al. (In prep)
Handika et. al. (2021)
Inclusive Open Source Software
Accessible software empowering new learners
From fieldwork to laboratory work, field-based evolutionary studies have become increasingly complex. For museum scientists, collecting specimens with dozens of accompanying parts are common practices nowadays. Genomic sequencing from thousands of loci to a whole genome have become the norm. I am working on developing open and accessible software to minimize the complexity, targeting the most neglected aspect of the workflow.
A language agnostic approach, cross-platforms, and extensive documentation
I utilize language agnostic approaches to produce efficient, reproducible, and inclusive software for field-based evolutionary studies. We take away the complexity from the users by simplifying installation process, reducing app dependencies, mobile and GUI support, and extensive documentation to aid new learners. We target wide-range of operating systems to ensure that our software is accessible to as many people as possible.
Project 1
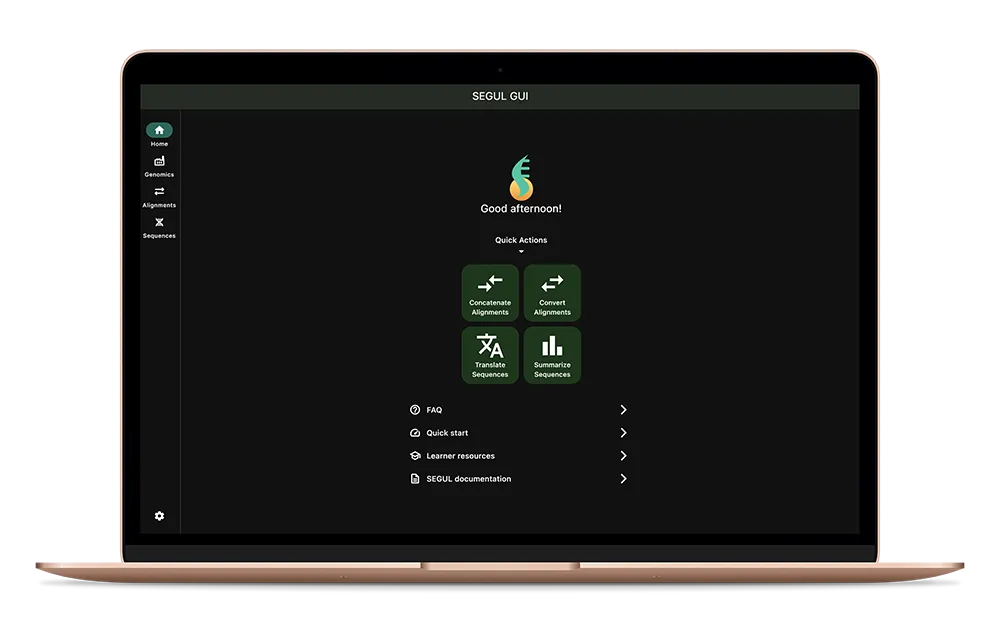
SEGUL/SEGUI, phylogenomic data manipulation and summary Statistics
The journey of my PhD has thought me that we are too reliant on High Performance Computers (HPC) to solve the complexity of phylogenomic studies. It includes the less computational intensive aspect, such as alignment manipulation and summary statistics. We rethink the approach to instead bringing phylogenomic to computers available for most people. SEGUL/SEGUI, is phylogenomic data manipulation and summary statistics software developed with the goal of being accessible to everyone. Of course, everyone means those who are interested in phylogenomics. 😊
SEGUL/SEGUI runs on everything. Almost literally. Starting from a single code base command line application written using Rust, we are able to scale SEGUL to support GUI interaction. It supports major platforms, from smartphone to HPC. We include extensive documentation to help beginners studying phylogenomics.
Publications:
Handika & Esselstyn (In review)
Project 2
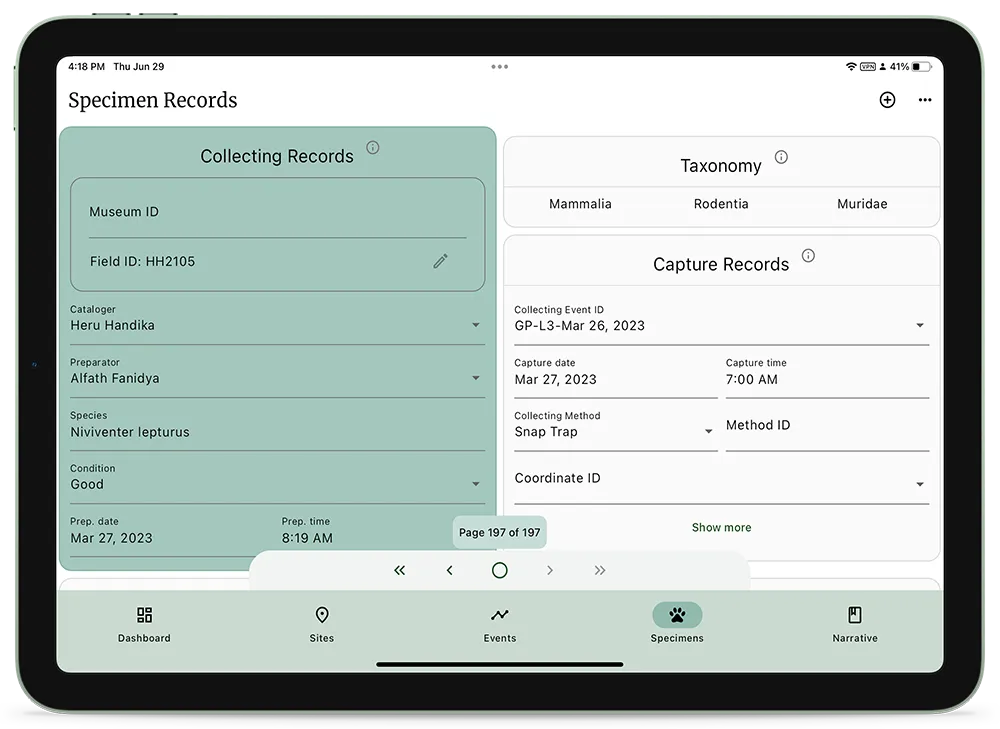
NAHPU, a field cataloging app for efficient data acquisition and improved collaboration
Nahpu is a collaborative effort to develop a field cataloging app to improve data acquisition efficiency and cross-institutional collaboration. Nahpu aims to minimize transcription errors, improved fieldwork reproducibility and data sharing, and provide a tool for everyone to conduct responsible field collecting.
Publications:
Handika et. al. (In Prep)
Other Projects
Specimen Digitization
This project is led by Dr. Nurainas at the Herbarium Universitas Andalas (ANDA) funded through the collection and data mobilization grant provided by the Global Biodiversity Information Facility (GBIF). Project progress is available here.
